MedShapeNet 2.0 Tutorial
MICCAI - 27th of September 2025 - 8.00-12.30
Welcome to our MICCAI (2025 - Daejeon) tutorial website for MedShapeNet 2.0. The tutorial will be hosted on the 27th of September, starting at 8.00!
Building: DCC1 Floor: F1 (ground floor), Room: 106.
Feel free to drop by anytime between 8.00-12.00
Here, you can find essential information about the program, speakers, organizers, and useful links:
- Program
- Speakers
- Organizers
- MedShapeNet Website
- Paper
- Preprint
- Python API (MedShapeNetCore) with Google Colab examples.
We invite you to join our tutorial. If you have any questions, please contact us.
MedShapeNet Overview
Before the deep learning era, statistical shape models (SSMs) were widely used in medical imaging. MedShapeNet builds upon this foundation, aiming to bridge computer vision methods to medical problems and clinical applications, inspired by benchmarks like ShapeNet and Princeton ModelNet.
MedShapeNet boasts a collection of over 100,000 medical shapes, covering bones, organs, vessels, muscles, and surgical instruments. These shapes can be searched, viewed in 3D, and downloaded individually using our shape search engine. It’s important to note that MedShapeNet is intended for research and educational purposes only. MedShapNet usefullness was demonstrated by its incorporation in research papers.
We will build MedShapeNet 2.0 with https://about.coscine.de/en/ as our database allowing for guaranteed 10+ years of hosting and deeper integration of our API.
Tutorial
The MedShapeNet 2.0 Tutorial will be held on 27 September 2025 (08:00–12:30) as part of MICCAI 2025.
This year’s theme is 3D shapes in medical imaging — highlighting methods of papers using MedShapeNet and complementary toolboxes.
The program brings together leading researchers who will present their recent papers, covering topics such as classification, generative models, point cloud analysis, registration, and shape toolkits. Some talks/papers are accompanied by code or datasets for participants to explore afterwards.
The tutorial emphasizes practical inspiration: rather than a step-by-step “installation/tutorial” session, methods and use cases are baked directly into the presentations. Discussion will be held online to maximize time and to connect participants directly with the speakers.
For details, see the speaker and program section.
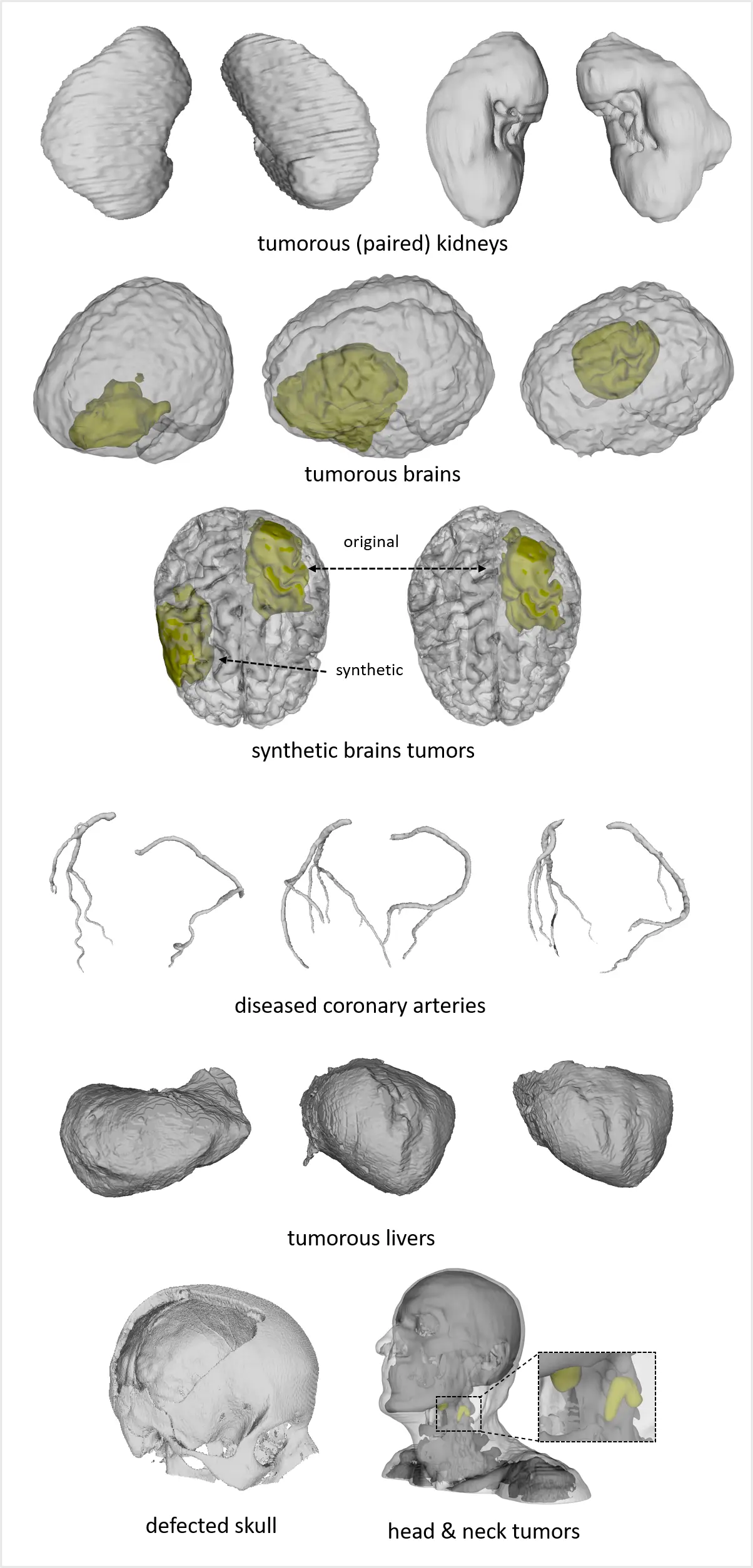
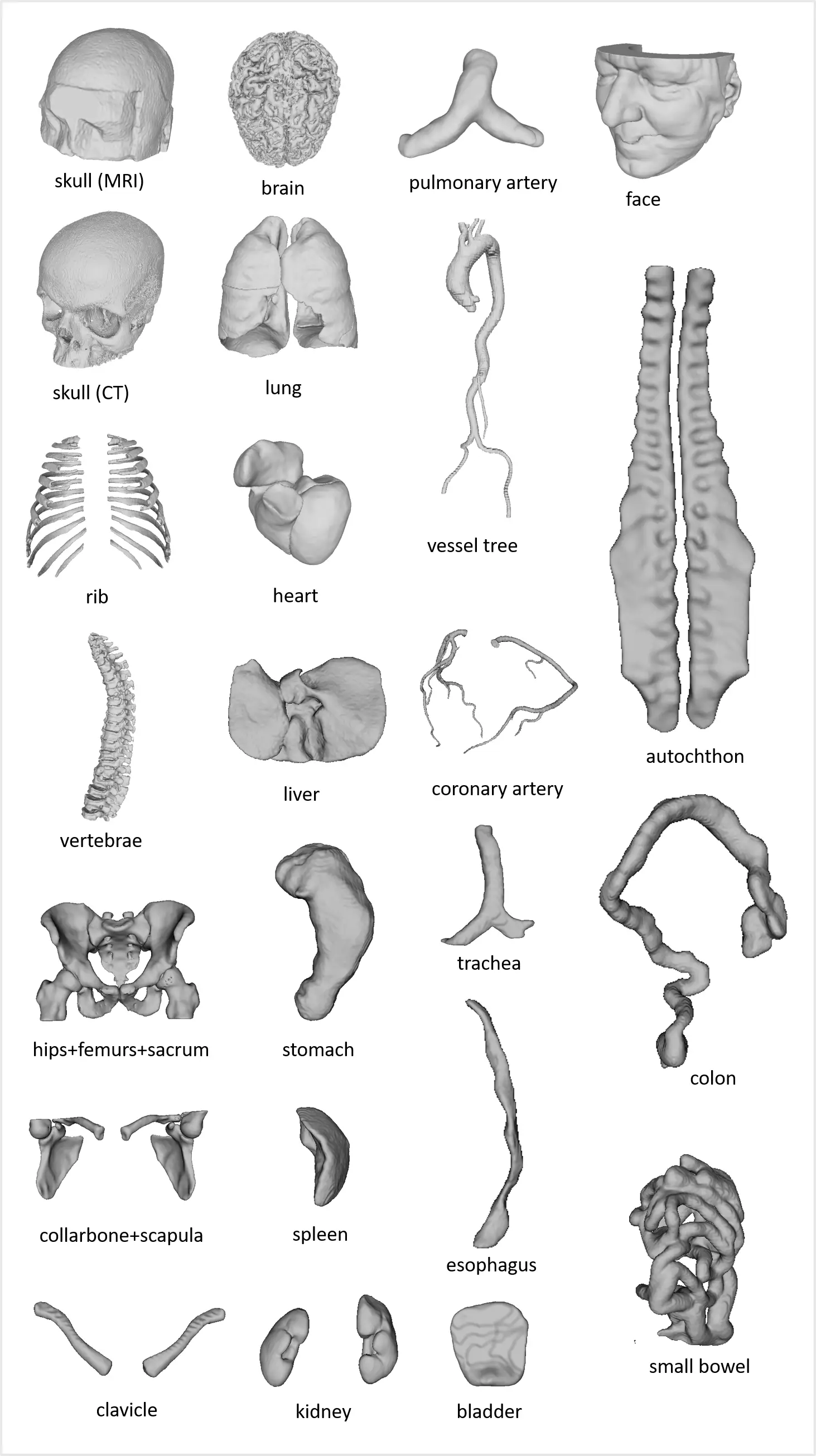
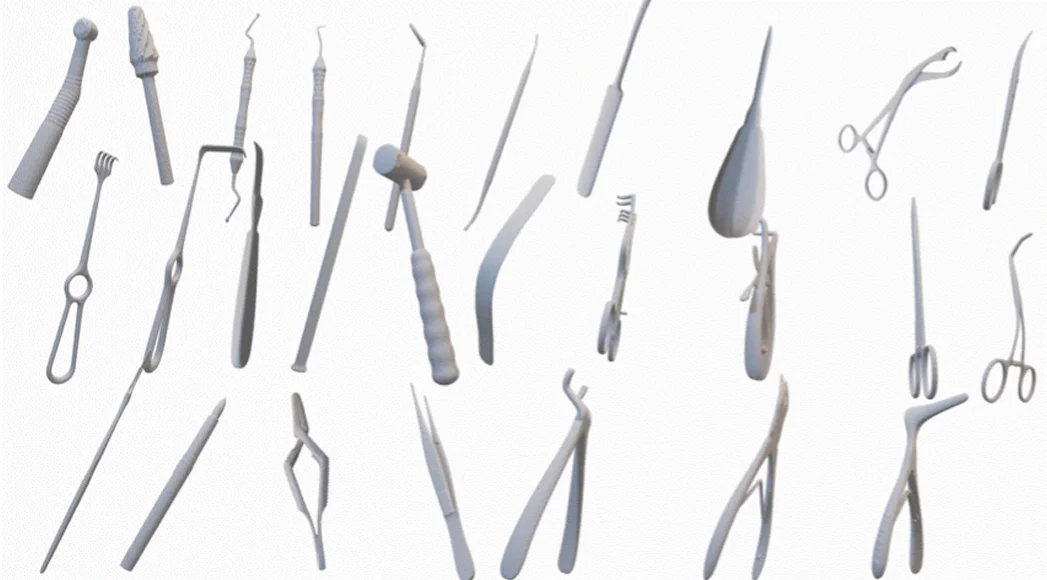
including anatomies (upper right), pathologies (upper left), and medical instruments (lower).
We refer to the MedShapeNet paper for more information on each individual dataset and its citation.
Speakers
Speakers have confirmed via email :-)
This year the focus will be on 3D Shapes in Medical imaging, toolboxes, and the methods of brilliant researchers utilizing MedShapeNet!
We have an amazing lineup of speakers - if possible, the discussion will be held online during the tutorial - maximizing our time and enabling getting in contact with the speaker of interest directly
Please note that these descriptions do not do them justice, please look up their respective profiles for more information
MSc. Gijs Luijten
M.Sc. Gijs Luijten, PhD candidate under Prof. Dr. Jan Egger at the AI-guided therapies group, works on AR/XR applications for treatment guidance and diagnosis. His background includes 3D scanning/printing and AR development at Radboudumc, with projects in Unity, Unreal Engine, and clinical XR datasets. He co-organizes workshops at MICCAI and is leading MedShapeNet, a large database of 3D medical shapes.
In this tutorial he will outline today’s program, highlight online resources, and guide the session.
More info: GitHub Samples/Showcases
Tomáš Krsička
MSc. Tomáš Krsička, master’s student at Brno University of Technology, converted his thesis on MedShapeNet into a published paper. His short talk will present a graph neural network autoencoder for 3D polygon meshes, introducing a novel pooling/unpooling operator, analysis of reconstruction performance, and the MedShapeNet19 dataset for benchmarking. Git repo NoteBook example
Dr. Titipat Achakulvisut & Dr. Peerapon Vateekul
Titipat Achakulvisut, Ph.D. in Bioengineering (UPenn), leads the Biomedical and Data Lab at Mahidol University. His research applies NLP and ML to biomedical data and scientific discovery, including recent work on skull reconstruction (IEEE Access, 2024). In this tutorial he will present methods from their IEEE paper.
Google Scholar
Peerapon Vateekul, Associate Professor at Chulalongkorn University, researches ML, deep learning, and NLP with medical projects including real-time polyp detection and gastroscopy segmentation. He has authored numerous works on classification, data mining, and medical AI, and will also discuss their IEEE paper.
Google Scholar
Guoqing Zhang
Tsinghua University researcher with a fresh preprint (April 2025) on Hierarchical Feature Learning for Medical Point Clouds via State Space Model. He introduces an SSM-based, multi-scale feature extractor for irregular medical point clouds, backed by a new dataset—MedPointS—tested on classification, completion, and segmentation tasks, and showing strong performance.
arXiv: 2504.13015 - Code!!!
Hantao Zhang
Researcher at EPFL. His recent work introduces a generative implicit medical shape model trained on MedShapeNet, enabling flexible generation and reconstruction of 3D shapes. This line of work builds on the MICCAI 2025 paper DiffAtlas (arXiv: 2503.06748) led by his supervisor Jiancheng Yang. Beyond generative modeling, he has also contributed to diffusion-based pathology synthesis and advanced segmentation methods presented at leading conferences and journals.
Julian Suk
Julian is a postdoctoral researcher at the Munich Center for Machine Learning (MCML) and Institute for AI and Informatics in Medicine at the Technical University of Munich (TUM). His research interest is geometric deep learning applied to cardiovascular hemodynamics modelling and cortical surface analysis. His latest preprint, GReAT (Aug 2025), shows how self-supervised models pretrained on 8,449 artery shapes improve wall shear stress prediction from just 49 clinical cases.
Francesca Maccarone on behalf of Prof. Simone Melzi
Prof. Simone Melzi, Associate Professor at University of Milano-Bicocca (DISCo), works at the intersection of geometry processing and AI. He is an ELLIS Scholar and recipient of the Eurographics Young Researcher Award 2023. With Maccarone, he co-authored S4A: Scalable Spectral Statistical Shape Analysis (STAG 2024), part of a suite of open-source, beginner-friendly shape-analysis projects. Francesca will deliver the talk on his behalf.
Yanghee Im
Yanghee Im is a PhD Student Researcher (Scholar) at the University of Southern California. She specializes in brain morphology and surface-based analysis. Author of A Scalable Toolkit for Modeling 3D Surface-based Brain Geometry, a toolkit that processes and analyzes multi-site data (N=3,373, 21 cohorts), enabling fine-grained subcortical mapping for psychiatric and neurodevelopmental research. Check out her poster at MICCAI as well! Paper!
Khoa Tuan Nguyen (tentative)
Medical imaging specialist with a focus on 3D shape in healthcare. Co-authored two recent MedShapeNet-related papers: one on advanced shape modeling (arXiv: 2508.02482) and another on complementary techniques (arXiv: 2504.19402), highlighting MedShapeNet’s growing impact. Additional info for the tutorial: (MICCAI paper, arXiv: 2508.02482, arXiv: 2504.19402)
Code related to the talk!
Wang Manning
Researcher at Fudan University, focusing on point-cloud shape registration and its application to computer-assisted interventions. His work explores how precise alignment of anatomical point clouds improves real-time guidance and planning.
Pedro Salvador Bassi
Developer of ShapeKit, a versatile toolkit designed to refine anatomical shapes—easy to integrate for researchers building shape-processing pipelines.
Zhe Min, Xinzhe Du & Shixing Ma
Team advancing registration methods in MedShapeNet. Zhe Min (UCL, Department of Medical Physics & Biomedical Engineering) has published widely on rigid and non-rigid registration, including multi-view 2D/3D alignment methods for computer-assisted surgery. Together with Du and Ma, their work enhances registration pipelines and interoperability across medical shape datasets.

Photo of last year's tutorial.
Program – MSN 2.0 Tutorial @ MICCAI 2025
The MSN 2.0 Tutorial will take place on 27 September 2025, 08:00–12:30, with a coffee break from 10:00–10:30.
This year’s theme is 3D shapes in medical imaging — highlighting toolboxes, methods, and papers that leverage MedShapeNet.
Location - Building: DCC1 - Floor F1 (ground floor) - Room: 106.
Speakers will share insights into classification, generative AI, point cloud analysis, and registration, with notebooks and datasets provided for participants to explore afterwards.
The discussion will be online (if possible with the wifi), allowing participants to directly connect with speakers of interest even after the event.
Resources and sample code can be found on our GitHub page.
Schedule
08:00 – 08:10
Opening & Program Overview
MSc. Gijs Luijten08:10 – 08:20
Tomáš Krsička
Benchmark-Ready 3D Anatomical Shape Classification and Dataset – with MSN
Short talk with notebook for participants. Git repo NoteBook example08:20 – 08:40
Dr. Titipat Achakulvisut
Skull Implants Generation and Categorization – with MSN
(IEEE Access 2024, IEEE Paper)08:40 – 09:00
Guoqing Zhang
Hierarchical Feature Learning for Medical Point Clouds via State Space Model – with MSN
arXiv: 2504.13015 - Code!!!09:00 – 09:20
Hantao Zhang
MedShapePrior: Towards a Generic Generative Implicit Shape Prior on MedShapeNet
A recent follow-up to DiffAtlas (MICCAI 2025) introduces a flexible generative framework for 3D shape reconstruction and synthesis. –> Code: GitHub09:20 – 09:35
Julian Suk
Leveraging Geometric Artery Data to Improve Wall Shear Stress Assessment – with MSN
Includes notebook and dataset.09:35 – 09:55
Prof. Simone Melzi / Francesca Maccarone
Shape Analysis and Overview of Projects
(Publications)
Papers and code that are inspiration for the talk: GeoFum Code Scalable spectral shape analysis - code - paper Unraveling Brainstem Deformations in Joubert Syndrome: A Statistical Shape Analysis of MRI-Derived Structures - Code - Paper Volumetric Functional Maps - Paper - Code09:55 – 10:30
Coffee Break10:30 – 10:45
Yanghee Im
Scalable Toolkit for Modeling 3D Surface-Based Brain Geometry10:45 – 11:05
Khoa Tuan Nguyen
From Generation to Evaluation: Building Better 3D Liver Shape Datasets.
(MICCAI paper, arXiv: 2508.02482, arXiv: 2504.19402)
Code related to the talk!11:05 – 11:25
Wang Manning
Point Cloud-Based Shape Registration and its Application in Computer-Assisted Intervention11:25 – 11:40
Pedro Salvador Bassi
ShapeKit – A Flexible Toolkit for Refining Anatomical Shapes11:40 – 12:10
Zhe Min, Xinzhe Du & Shixing Ma
MedShapeNet and Registration
Advances in rigid/non-rigid registration and interoperability for medical shapes.12:10 – 12:15
Closing & Outlook
MSc. Gijs Luijten
The following author is not able to present but was so kind to contribute a short talk, notebook, code and more: Laverde, N., Robles, M., & Rodríguez, J. (2025). Shap-MeD. arXiv:2503.15562. Universidad de los Andes. https://arxiv.org/abs/2503.15562
3-minute video explaining the project and including a demo Git repository with all the code The Hugging Face repository containing both the dataset and the trained model
If you would like to run the trained model directly, you can find the detailed steps in this section of the README with a test notebook
Note: Each slot includes ~1–2 minutes for questions and transition. The schedule is designed to fit exactly within the 08:00–12:30 session. Further discussion is promoted online and/or after the event.
Notes
- Total presentation time: 240 minutes, including break.
- Short buffers allow for 1–2 questions per speaker - participants are encouraged to write/contact our speakers.
Just to let you know, participants are welcome to join at any time during the tutorial; don’t be shy (in case you miss a bus or want to drop by shortly).
Schedule (Table Overview)
| Time | Speaker | Title |
|---|---|---|
| 08:00–08:10 | MSc. Gijs Luijten | Opening & Program Overview |
| 08:10–08:20 | Tomáš Krsička | Benchmark-Ready 3D Anatomical Shape Classification and Dataset – MSN |
| 08:20–08:40 | Dr. Titipat Achakulvisut | Skull Implants Generation and Categorization – MSN (IEEE Access 2024, IEEE Paper) |
| 08:40–09:00 | Guoqing Zhang | Hierarchical Feature Learning for Medical Point Clouds via SSM – MSN (arXiv: 2504.13015) |
| 09:00–09:20 | Hantao Zhang | Generative Implicit Medical Shape Model on MedShapeNet – follow-up to DiffAtlas (MICCAI 2025) |
| 09:20–09:35 | Julian Suk | Leveraging Geometric Artery Data to Improve Wall Shear Stress Assessment – MSN |
| 09:35–09:55 | Prof. Simone Melzi / F. Maccarone | Shape Analysis and Overview of Projects (Publications) |
| 09:55–10:30 | — | Coffee Break |
| 10:30–10:45 | Yanghee Im | Scalable Toolkit for Modeling 3D Surface-Based Brain Geometry |
| 10:45–11:05 | Khoa Tuan Nguyen (tentative) | From Generation to Evaluation: Building Better 3D Liver Shape Datasets. (MICCAI paper, Code related to the talk! |
| 11:05–11:25 | Wang Manning | Point Cloud-Based Shape Registration for Computer-Assisted Intervention |
| 11:25–11:40 | Pedro Salvador Bassi | ShapeKit – A Flexible Toolkit for Refining Anatomical Shapes |
| 11:40–12:10 | Zhe Min, Xinzhe Du & Shixing Ma | MedShapeNet and Registration |
| 12:10–12:15 | MSc. Gijs Luijten | Closing & Outlook |
The following author is not able to present but was so kind to contribute a short talk, notebook, code and more: Laverde, N., Robles, M., & Rodríguez, J. (2025). Shap-MeD. arXiv:2503.15562. Universidad de los Andes. https://arxiv.org/abs/2503.15562
3-minute video explaining the project and including a demo Git repository with all the code The Hugging Face repository containing both the dataset and the trained model
If you would like to run the trained model directly, you can find the detailed steps in this section of the README with a test notebook
Note: Each slot includes ~1–2 minutes for questions and transition. The schedule is designed to fit exactly within the 08:00–12:30 session.
Python Toolbox (API)
for 3D Medical Shape Analysis
MedShapeNet 2.0 API is the continuation of MedShapeNet and includes all functionality of MedShapeNet 1.0 and MedShapeNetCore.
MedShapeNet 2.0 strives towards a AWS compliant S3 COSCINE storage which will be funded by NRW and host the data for a minimum of ten years.
We hope to demonstrate the improved MedShapeNet and enable researchers to not only contribute datasets, but also showcases, code and functionality.
FYI: MedShapeNetCore is a subset of MedShapeNet, containing more lightweight 3D anatomical shapes in the format of mask, point cloud and mesh.
The shape data are stored as numpy arrays in nested dictonaries in npz format Zenodo.
This API provides means to downloading, accessing and processing the shape data via Python, which integrates MedShapeNetCore seamless into Python-based machine learning workflows.
Contact
Master students are encouraged to mail when interested in an internship.
- Gijs.Luijten@uk-essen.de
- Institute for AI in Medicine, Giradet Straße 2, 45131, Essen, Germany
- IKIM-UME